16.2 Remote data module
Remote data module retrieves remote sensing data from MODISTools, Google Earth Engine and AppEEARS, as well as from the NASA’s Land Processes Distributed Active Archive Center (LP DAAC) data server. For currently available R functions see here, and for the Python functions see RpTools. The downloaded data can be used while performing further analyses in PEcAn.
16.2.0.1 Google Earth Engine
Google Earth Engine is a cloud-based platform for performing analysis on satellite data. It provides access to a large data catalog through an online JavaScript code editor and a Python API.
Datasets currently available for use in PEcAn via Google Earth Engine are,
- Sentinel-2 MSI
gee2pecan_s2(). It is possible to estimate Leaf Area Index (LAI) from Sentinel-2 data using the ESA SNAP algorithm. - SMAP Global Soil Moisture Data
gee2pecan_smap() - Landsat 8 Surface Reflectance
gee2pecan_l8() - Global Forest Canopy Height, 2019
gee2pecan_gedi()
16.2.0.2 AppEEARS
AppEEARS (Application for Extracting and Exploring Analysis Ready Samples) is an online tool which provides an easy to use interface for downloading analysis ready remote sensing data. Products available on AppEEARS. Note: AppEEARS uses a task based system for processing the data request, it is possible for a task to run for long hours before it gets completed. The module checks the task status after every 60 seconds and saves the files when the task gets completed.
16.2.0.3 LP DAAC Data Pool
LP DAAC (Land Processes Distributed Active Archive Center) Data Pool is a NASA Earthdata login-enabled server located at the USGS Earth Resources Observation and Science (EROS) Center that archives and distributes land data products. Similar to AppEEARS, using this source also requires an Earthdata account (see below). Currently this pipeline is implemented and tested only for the GEDI dataset. For more information about the data you are downloading, including documentation and how to properly cite the data, please visit https://lpdaac.usgs.gov/.
16.2.0.4 Set-Up instructions (first time and one time only):
Sign up for the Google Earth Engine. Follow the instructions here to sign up for using GEE. You need to have your own GEE account for using the GEE download functions in this module.
Sign up for NASA Earthdata. Using AppEEARS and LP DAAC Data Pool requires an Earthdata account visit this page to create your own account.
Install the RpTools package. Python codes required by this module are stored in a Python package named “RpTools” using this requires Python3 and the package manager pip3 to be installed in your system. To install the package,
- Navigate to
pecan/modules/data.remote/inst/RpToolsIf you are inside the pecan directory, this can be done by,
- Use pip3 to install the package. “-e” flag is used to install the package in an editable or develop mode, so that changes made to the code get updated in the package without reinstalling.
- Authenticate GEE. The GEE API needs to be authenticated using your credentials. The credentials will be stored locally on your system. This can be done by,
#this will open a browser and ask you to sign in with the Google account registered for GEE
earthengine authenticateAlternate way,
- Save the Earthdata credentials. If you wish to use AppEEARS or LP DAAC data pool you will have to store your username and password inside a JSON file and then pass its file path as an argument in
remote_process
16.2.0.5 Usage guide:
This module is accesible using the R function remote_process which uses the Python package “RpTools” (located at data.remote/inst/RpTools) for downloading and processing data. RpTools has a function named rp_control which controls two other functions,
get_remote_datawhich controls the scripts which are used for downloading data from the source. For example,gee2pecan_s2downloads bands from Sentinel 2 using GEE.process_remote_datawhich controls the scripts responsible for processing the raw data. For example,bands2lai_snapuses the downloaded bands to compute LAI using the SNAP algorithm.
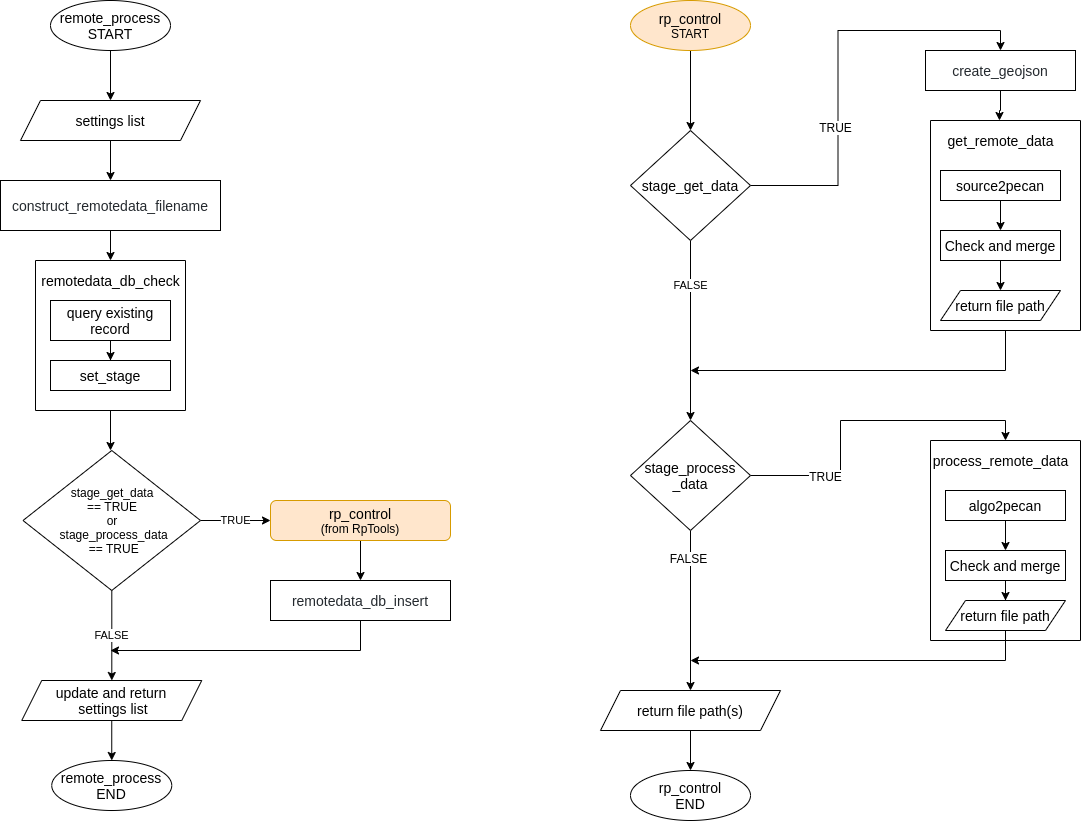
16.2.1 Configuring remote_process
remote_process is configured using remote data tags in the pecan.xml. The required tags are described below,
<remotedata>
<out_get_data>...</out_get_data>
<source>...</source>
<collection>...</collection>
<scale>...</scale>
<projection>...</projection>
<qc>...</qc>
<algorithm>...</algorithm>
<credfile>...</credfile>
<out_process_data>...</out_process_data>
<overwrite>...</overwrite>
</remotedata>out_get_data: (required) type of raw output requested, e.g, bands, smapsource: (required) source of remote data, e.g., gee or appeearscollection: (required) dataset or product name as it is provided on the source, e.g. “COPERNICUS/S2_SR” for gee or “SPL3SMP_E.003” for appeearsscale: (optional) pixel resolution required for some gee collections, recommended to use 10 for Sentinel 2 scale Information about how GEE handles scale can be found out hereprojection: (optional) type of projection. Only required for appeears polygon AOI typeqc: (optional) quality control parameter, required for some gee collectionsoverwrite: (optional) if TRUE database checks will be skipped and existing data of same type will be replaced entirely. When processed data is requested, the raw data required for creating it will also be replaced. By default FALSE
If you don’t want to enter your Earthdata credentials everytime you use AppEEARS or LP DAAC use the following tag too:
credfile: (optional) absolute path to JSON file containing Earthdata username and password, only required when using AppEEARS and LP DAAC data pool. The contents of this file could be as simple as the following:
{
"username": "yourEARTHDATAusername",
"password": "yourEARTHDATApassword"
}These tags are only required if processed data (i.e. further processing after downloading the data) is requested:
out_process_data: (optional) type of processed output requested, e.g, laialgorithm: (optional) algorithm used for processing data, currently only SNAP is implemented to estimate LAI from Sentinel-2 bands
Additional information are taken from the registration files located at pecan/modules/data.remote/inst/registration, each source has its own registration file. This is so because there isn’t a standardized way to retrieve all image collections from GEE and each image collection may require its own way of performing quality checks, etc whereas all of the products available on AppEEARS can be retrieved using its API in a standardized way.
GEE registration file (register.GEE.xml) :
collectionoriginal_nameoriginal name of the image collection, e.g. COPERNICUS/S2_SRpecan_nameshort form of original name using which the collection is represented in PEcAn, e.g. s2
coordcoord_typecoordinate type supported by the collection
scalethe default value of the scale can be specified hereqcthe default value of the qc parameter can be specified hereraw_formatformat details of the raw fileidid of the formatnamename of the formatmimetypeMIME type
pro_formatformat details of the processed file when the collection is used to create a processed fileidid of the formatnamename of the formatmimetypeMIME type
AppEEARS and LP DAAC Data Pool registration files (register.APPEEARS.xml and register.LPDAACDATAPOOL.xml) :
coordcoord_typecoordinate type supported by the product
raw_formatformat details of the output fileidid of the formatnamename of the formatmimetypeMIME type
Remaining input data:
- start date, end date: these are taken from the
runtag inpecan.xml - outdir: from the
outdirtag inpecan.xml - Area of interest: the coordinates and site name are found out from BETY using
siteidpresent in theruntag. These are then used to create a GeoJSON file which is used by the download functions.
The output data from the module are returned in the following tags:
raw_id: input id of the raw fileraw_path: absolute path to the raw filepro_id: input id of the processed filepro_path: absolute path to the processed file
Output files:
The output files are of netCDF type and are stored in a directory inside the specified outdir with the following naming convention: source_site_siteid
The output files are created with the following naming convention: source_collection_scale_projection_qc_site_siteid_TimeStampOfFileCreation
Whenever a data product is requested the output files are stored in the inputs table of BETYdb. Subsequently when the same product is requested again with a different date range but with the same qc, scale, projection the previous file in the db would be extended. The DB would always contain only one file of the same type. As an example, if a file containing Sentinel 2 bands for start date: 2018-01-01, end date: 2018-06-30 exists in the DB and the same product is requested again for a different date range one of the following cases would happen,
New dates are ahead of the existing file: For example, if the requested dates are start: 2018-10-01, end: 2018-12-31 in this case the previous file will be extended forward meaning the effective start date of the file to be downloaded would be the day after the end date of the previous file record, i.e. 2018-07-01. The new and the previous file would be merged and the DB would now be having data for 2018-01-01 to 2018-12-31.
New dates are preceding of the existing file: For example, if the requested dates are start: 2017-01-01, end: 2017-06-30 in this case the effective end date of the new download would be the day before the start date of the existing file, i.e., 2017-12-31. The new and the previous file would be merged and the file in the DB would now be having data for 2017-01-01 to 2018-06-30.
New dates contain the date range of the existing file: For example, if the requested dates are start: 2016-01-01, end: 2019-06-30 here the existing file would be replaced entirely with the new file. A more efficient way of doing this could be to divide your request into two parts, i.e, first request for 2016-01-01 to 2018-01-01 and then for 2018-06-30 to 2019-06-30.
When a processed data product such as SNAP-LAI is requested, the raw product (here Sentinel 2 bands) used to create it would also be stored in the DB. If the raw product required for creating the processed product already exists for the requested time period, the processed product would be created for the entire time period of the raw file. For example, if Sentinel 2 bands are present in the DB for 2017-01-01 to 2017-12-31 and SNAP-LAI is requested for 2017-03-01 to 2017-07-31, the output file would be containing LAI for 2017-01-01 to 2017-12-31.
16.2.1.1 Creating Polygon based sites
A polygon site can be created in the BETYdb using the following way,
PEcAn.DB::db.query("insert into sites (country, sitename, geometry) values ('country_name', 'site_name', ST_SetSRID(ST_MakePolygon(ST_GeomFromText('LINESTRING(lon lat elevation)')), crs));", con)Example,
db.query("insert into sites (country, sitename, geometry) values ('FI', 'Qvidja_ca6cm', ST_SetSRID(ST_MakePolygon(ST_GeomFromText('LINESTRING(22.388957339620813 60.287395608412218 14.503780364990234, 22.389600591651835 60.287182336733203 14.503780364990234,
22.38705422266651 60.285516177775868 14.503780364990234,
22.386575219445195 60.285763643883932 14.503780364990234,
22.388957339620813 60.287395608412218 14.503780364990234 )')), 4326));", con)16.2.1.2 Example use (GEE)
This example will download Sentinel 2 bands and then use the SNAP algorithm to compute Leaf Area Index.
- Add remotedata tag to
pecan.xmland configure it.
<remotedata>
<out_get_data>bands</out_get_data>
<source>gee</source>
<collection>COPERNICUS/S2_SR</collection>
<scale>10</scale>
<qc>1</qc>
<algorithm>snap</algorithm>
<out_process_data>LAI</out_process_data>
</remotedata>- Store the contents of
pecan.xmlin a variable namedsettingsand pass it toremote_process.
PEcAn.data.remote::remote_process(settings)The output netCDF files(bands and LAI) will be saved at outdir and their records would be kept in the inputs table of BETYdb.
16.2.1.3 Example use (AppEEARS)
This example will download the layers of a SMAP product(SPL3SMP_E.003)
- Add remotedata tag to
pecan.xmland configure it.
<remotedata>
<out_get_data>smap</out_get_data>
<source>appeears</source>
<collection>SPL3SMP_E.003</collection>
<projection>native</projection>
<algorithm></algorithm>
<credfile>path/to/jsonfile/containingcredentials</credfile>
</remotedata>- Store the contents of
pecan.xmlin a variable namedsettingsand pass it toremote_process.
PEcAn.data.remote::remote_process(settings)The output netCDF file will be saved at outdir and its record would be kept in the inputs table of BETYdb.
16.2.1.4 Example use GEDI (LP DAAC data pool)
<remotedata>
<out_get_data>gedi</out_get_data>
<source>lpdaacdatapool</source>
<collection>GEDI02_B.002</collection>
<credfile>path/to/jsonfile/containingcredentials</credfile>
</remotedata>- Store the contents of
pecan.xmlin a variable namedsettingsand pass it toremote_process.
PEcAn.data.remote::remote_process(settings)16.2.1.5 Adding new GEE image collections
Once you have the Python script for downloading the collection from GEE, please do the following to integrate it with this module.
Make sure that the function and script names are same and named in the following way:
gee2pecan_pecancodeofimagecollectionpecancodeofimagecollectioncan be any name which you want to use for representing the collection is an easier way. Additionaly, ensure that the function accepts and uses the following arguments,geofile- (str) GeoJSON file containing AOI information of the siteoutdir- (str) path where the output file has to be savedstart- (str) start date in the form YYYY-MM-DDend- (str) end date in the form YYYY-MM-DDscaleandqcif applicable.
Make sure the output file is of netCDF type and follows the naming convention described above.
Store the Python script at
pecan/modules/data.remote/inst/RpTools/RpToolsUpdate the
register.GEE.xmlfile.
After performing these steps the script will be integrated with the remote data module and would be ready to use.