18.2 Example met conversion wrapper function
met2model.MODEL <- function(in.path, in.prefix, outfolder, start_date, end_date){ myMetScript <- system.file(“inst/met2model.MODEL.sh”, “PEcAn.MODEL”) system(paste(myMetScript, file.path(in.path, in.prefix), outfolder, start_date, end_date)) }
would execute the following at the Linux command line
inst/met2model.MODEL.sh in.path/in.prefix outfolder start_date end_date `
18.2.0.1 DESCRIPTION
Within the module folder open the DESCRIPTION file and change the package name to PEcAn.MODEL. Fill out other fields such as Title, Author, Maintainer, and Date.
18.2.0.2 NAMESPACE
This file is managed by Roxygen and will update automatically when you build the model package. Do not edit it by hand.
18.2.0.3 Building the package
if you have a favorite tool for building R packages from local directories, you can use it as normal during development. If you do not yet have a favorite, use PEcAn’s Make system: Add your package to the Makefile by adding its name to the line starting MODELS := near the top of the file. Then you can update Roxygen output with make document, build and install the package with make install, and run package checks and tests with make check / make check. Since these run on every PEcAn package by default, you may want to limit these to the MODEL package with make .check/models/MODEL.
18.2.0.4 write.config.MODEL (required)
This module performs two primary tasks. The first is to take the list of parameter values and model input files that it receives as inputs and write those out in whatever format(s) the MODEL reads (e.g. a settings file). The second is to write out a shell script, jobs.sh, which, when run, will start your model run and convert its output to the PEcAn standard (netCDF with metadata currently equivalent to the MsTMIP standard). Within the MODEL directory take a close look at inst/template.job and the example write.config.MODEL to see an example of how this is done. It is important that this script writes or moves outputs to the correct location so that PEcAn can find them. The example function also shows an example of writing a model-specific settings/config file, also by using a template.
You are encouraged to read the section below on defining PFTs before writing write.config.MODEL so that you understand what model parameters PEcAn will be passing you, how they will be named, and what units they will be in. Also note that the (optional) PEcAn input/driver processing scripts are called by separate workflows, so the paths to any required inputs (e.g. meteorology) will already be in the model-specific format by the time write.config.MODEL receives that info.
18.2.0.5 Output Conversions
The module model2netcdf.MODEL converts model output into the PEcAn standard (netCDF with metadata currently equivalent to the MsTMIP standard). This function was previously required, but now that the conversion is called within jobs.sh it may be easier for you to convert outputs using other approaches (or to just directly write outputs in the standard).
Whether you implement this function or convert outputs some other way, please note that PEcAn expects all outputs to be broken up into ANNUAL files with the year number as the file name (i.e. YEAR.nc), though these files may contain any number of scalars, vectors, matrices, or arrays of model outputs, such as time-series of each output variable at the model’s native timestep.
Note: PEcAn reads all variable names from the files themselves so it is possible to add additional variables that are not part of the MsTMIP standard. Similarly, there are no REQUIRED output variables, though time is highly encouraged. We are shortly going establish a canonical list of PEcAn variables so that if users add additional output variables they become part of the standard. We don’t want two different models to call the same output with two different names or different units as this would prohibit the multi-model syntheses and comparisons that PEcAn is designed to facilitate.
18.2.0.6 met2model.MODEL
met2model.MODEL(in.path, in.prefix, outfolder, start_date, end_date)
Converts meteorology input files from the PEcAn standard (netCDF, CF metadata) to the format required by the model. This file is optional if you want to load all of your met files into the Inputs table as described in How to insert new Input data, which is often the easiest way to get up and running quickly. However, this function is required if you want to benefit from PEcAn’s meteorology workflows and model run cloning. You’ll want to take a close look at [Adding-an-Input-Converter] to see the exact variable names and units that PEcAn will be providing. Also note that PEcAn splits all meteorology up into ANNUAL files, with the year number explicitly included in the file name, and thus what PEcAn will actually be providing is in.path, the input path to the folder where multiple met files may stored, and in.prefix, the start of the filename that precedes the year (i.e. an individual file will be named <in.prefix>.YEAR.nc). It is valid for in.prefix to be blank. The additional REQUIRED arguments to met2model.MODEL are outfolder, the output folder where PEcAn wants you to write your meteorology, and start_date and end_date, the time range the user has asked the meteorology to be processed for.
18.2.0.7 Additional Input Converters
In addition to met2model.MODEL, PEcAn also supports the following additional input conversions: * veg2model.MODEL - handles vegetation initial conditions. Supports both pool-based and cohort-based models * write.events.MODEL - handles management events (still in early development) * soil physical parameters (e.g., texture, hydraulics, thermodynamics) - there is not yet a convention for stand-alone converters, existing models currently read the soil.nc file from settings\(run\)inputs\(soil_physics as part of their write.configs. This file is generated by PEcAn.data.land::soil2netcdf * vegetation phenology - there is not yet a convention for stand-alone converters, existing models read the phenology file from settings\)run\(inputs\)leaf_phenology within their write.configs. This is currently a csv file with the columns: “year”, “site_id”, “lat”, “lon”, “leafonday”,“leafoffday”,“leafon_qa”,“leafoff_qa”
Because not all models accept these inputs, the pecan.MODEL package template does not include converter skeletons for them. See the code of existing model coupler packages to find examples, and ask freely in Slack for suggestions specific to your project.
See also Adding a new input converter for more about PEcAn’s approach to input handling and what information is passed in each input type.
18.2.0.8 Commit changes
Once the MODEL modules are written, you should follow the Using-Git instructions on how to commit your changes to your local git repository, verify that PEcAn compiles using scripts/build.sh, push these changes to Github, and submit a pull request so that your model module is added to the PEcAn system. It is important to note that while we encourage users to make their models open, adding the PEcAn interface module to the Github repository in no way requires that the model code itself be made public. It does, however, allow anyone who already has a copy of the model code to use PEcAn so we strongly encourage that any new model modules be committed to Github.
18.2.1 Integrate MODEL into the PEcAn build system
Once the package is defined, you will also want to make the rest of PEcAn notice it:
- Add it to the PEcAn Makefile, as part of the line near the top that starts
MODELS :=(if you didn’t do this earlier to build the package). - Run
scripts/generate_dependencies.Rto add the list of your model’s dependencies to the packages pre-installed on the PEcAn docker images. - Add it to the list of packages in
base/all/data/pecan_version_history.csv - Optionally, add it as a
Suggests:dependency in base/all/DESCRIPTION (not all models choose to do this). - If your model package has a Dockerfile, add it to the
modelsbinarysection of the Docker build in `.github/workflows/docker.yml - Once the package is working and merged into the develop branch of PEcAn, open a pull request to add it to the
packages.jsonfile of [https://github.com/PecanProject/pecanproject.r-universe.dev]. This will cue R-Universe to start building and distributing the package as part of the PEcAn collection. - Announce it in the CHANGELOG!
18.2.2 Add model info to PEcAn Database
Note: As support for running PEcAn with no database expands, this step has become less important, but keep reading – this section is still the best summary of what information needs to be available to the model, whether looked up from the database on the fly or passed in by hand.
To run a model within PEcAn requires that the PEcAn database has sufficient information about the model. This includes a MODEL_TYPE designation, the types of inputs the model requires, the location of the model executable, and the plant functional types used by the model.
The instructions in this section assume that you will be specifying this information using the BETYdb web-based interface. This can be done either on your local VM (localhost:3280/bety or localhost:6480/bety) or on a server installation of BETYdb. However you interact with BETYdb, we encourage you to set up your PEcAn instance to support database syncs so that these changes can be shared and backed-up across the PEcAn network.
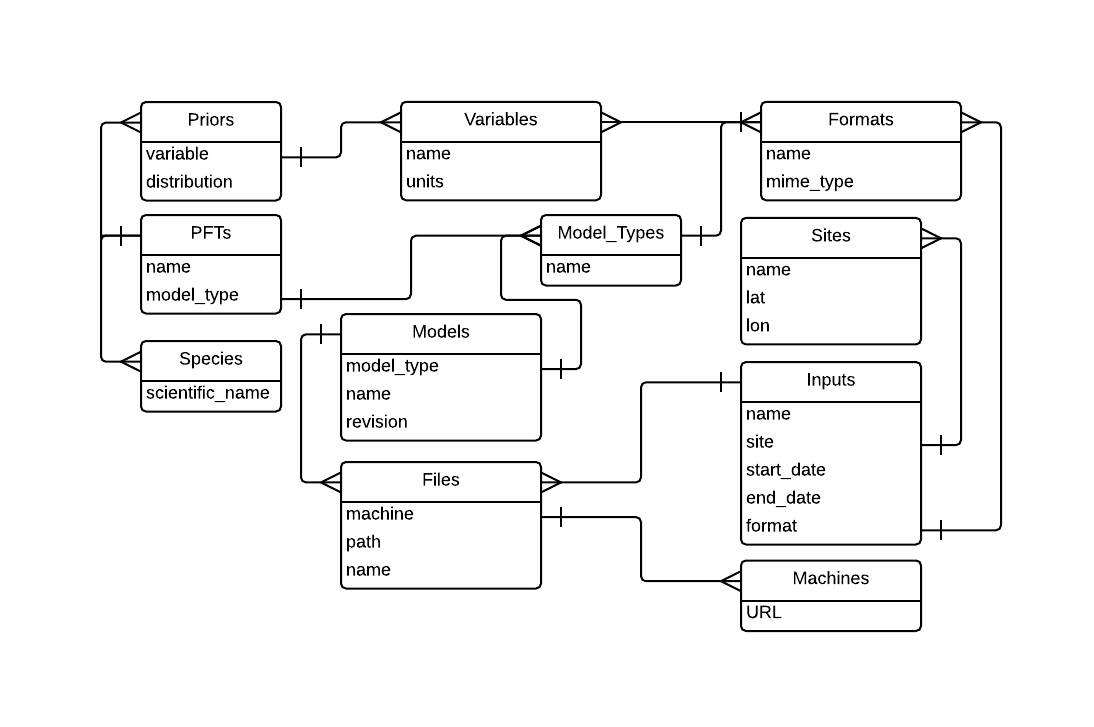
The figure below summarizes the relevant database tables that need to be updated to add a new model and the primary variables that define each table.
18.2.3 Define MODEL_TYPE
The first step to adding a model is to create a new MODEL_TYPE, which defines the abstract model class. This MODEL_TYPE is used to specify input requirements, define plant functional types, and keep track of different model versions.
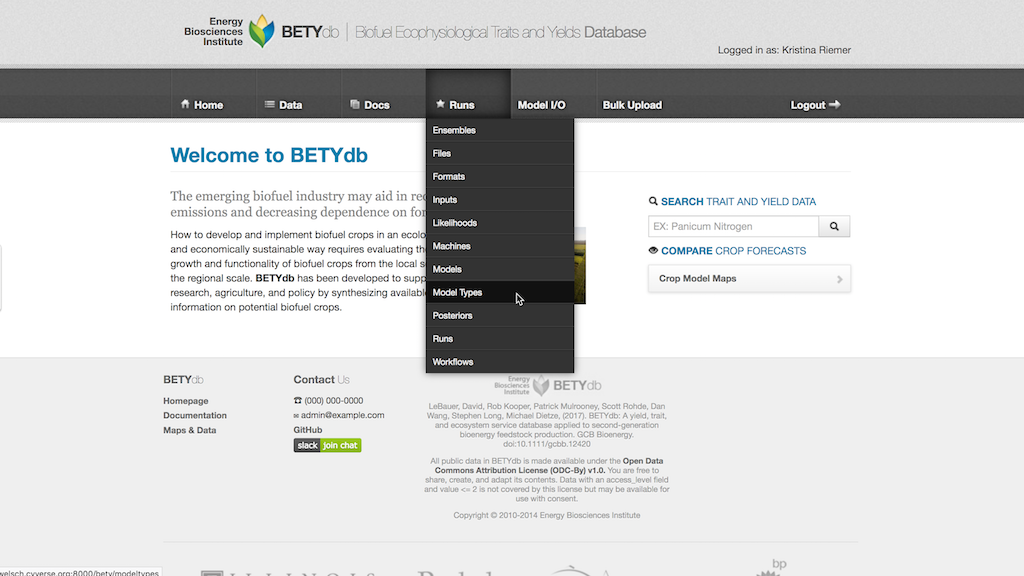
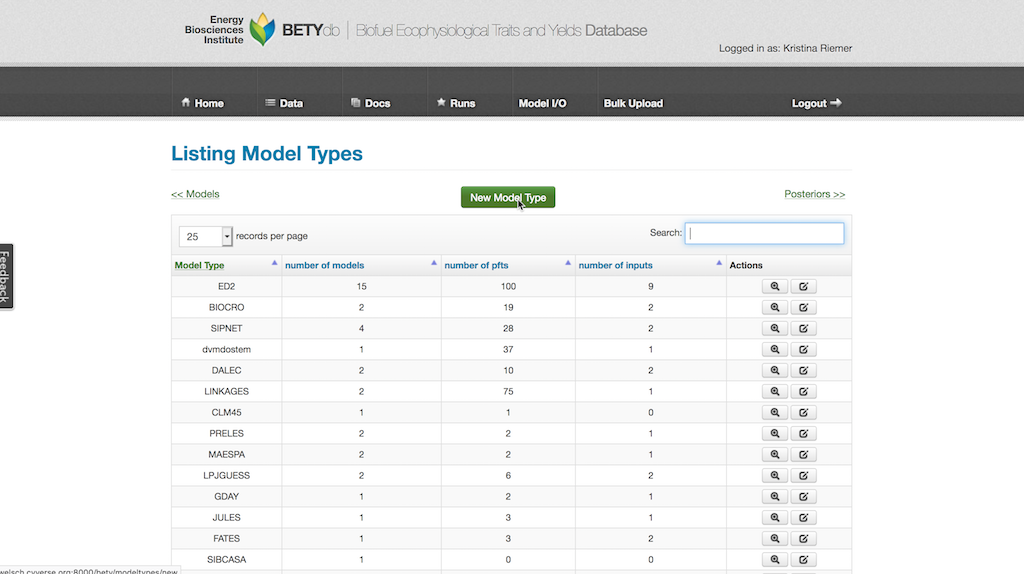
The MODEL_TYPE is created by selecting Runs > Model Type and then clicking on New Model Type. The MODEL_TYPE name should be identical to the MODEL package name (see Interface Module below) and is case sensitive.
18.2.4 MACHINE
The PEcAn design acknowledges that the same model executables and input files may exist on multiple computers. Therefore, we need to define the machine that that we are using. If you are running on the VM then the local machine is already defined as pecan. Otherwise, you will need to select Runs > Machines, click New Machine, and enter the URL of your server (e.g. pecan2.bu.edu).
18.2.5 MODEL
Next we are going to tell PEcAn where the model executable is. Select Runs > Files, and click ADD. Use the pull down menu to specify the machine you just defined above and fill in the path and name for the executable. For example, if SIPNET is installed at /usr/local/bin/sipnet then the path is /usr/local/bin/ and the file (executable) is sipnet.
Now we will create the model record and associate this with the File we just registered. The first time you do this select Runs > Models and click New Model. Specify a descriptive name of the model (which doesn’t have to be the same as MODEL_TYPE), select the MODEL_TYPE from the pull down, and provide a revision identifier for the model (e.g. v3.2.1). Once the record is created select it from the Models table and click EDIT RECORD. Click on “View Related Files” and when the search window appears search for the model executable you just added (if you are unsure which file to choose you can go back to the Files menu and look up the unique ID number). You can then associate this Model record with the File by clicking on the +/- symbol. By contrast, clicking on the name itself will take you to the File record.
In the future, if you set up the SAME MODEL VERSION on a different computer you can add that Machine and File to PEcAn and then associate this new File with this same Model record. A single version of a model should only be entered into PEcAn once.
If a new version of the model is developed that is derived from the current version you should add this as a new Model record but with the same MODEL_TYPE as the original. Furthermore, you should set the previous version of the model as Parent of this new version.
18.2.6 FORMATS
The PEcAn database keep track of all the input files passed to models, as well as any data used in model validation or data assimilation. Before we start to register these files with PEcAn we need to define the format these files will be in. To create a new format see Formats Documentation.
18.2.7 MODEL_TYPE -> Formats
For each of the input formats you specify for your model, you will need to edit your MODEL_TYPE record to add an association between the format and the MODEL_TYPE. Go to Runs > Model Type, select your record and click on the Edit button. Next, click on “Edit Associated Formats” and choose the Format you just defined from the pull down menu. If the Input box is checked then all matching Input records will be displayed in the PEcAn site run selection page when you are defining a model run. In other words, the set of model inputs available through the PEcAn web interface is model-specific and dynamically generated from the associations between MODEL_TYPEs and Formats. If you also check the Required box, then the Input will be treated as required and PEcAn will not run the model if that input is not available. Furthermore, on the site selection webpage, PEcAn will filter the available sites and only display pins on the Google Map for sites that have a full set of required inputs (or where those inputs could be generated using PEcAn’s workflows). Similarly, to make a site appear on the Google Map, all you need to do is specify Inputs, as described in the next section, and the point should automatically appear on the map.
18.2.8 INPUTS
After a file Format has been created then input files can be registered with the database. Creating Inputs can be found under How to insert new Input data.
18.2.9 Add Plant Functional Types (PFTs)
Since many of the PEcAn tools are designed to keep track of parameter uncertainties and assimilate data into models, to use PEcAn with a model it is important to define Plant Functional Types for the sites or regions that you will be running the model.
Create a new PFT entry by selecting Data > PFTs and then clicking on New PFT.
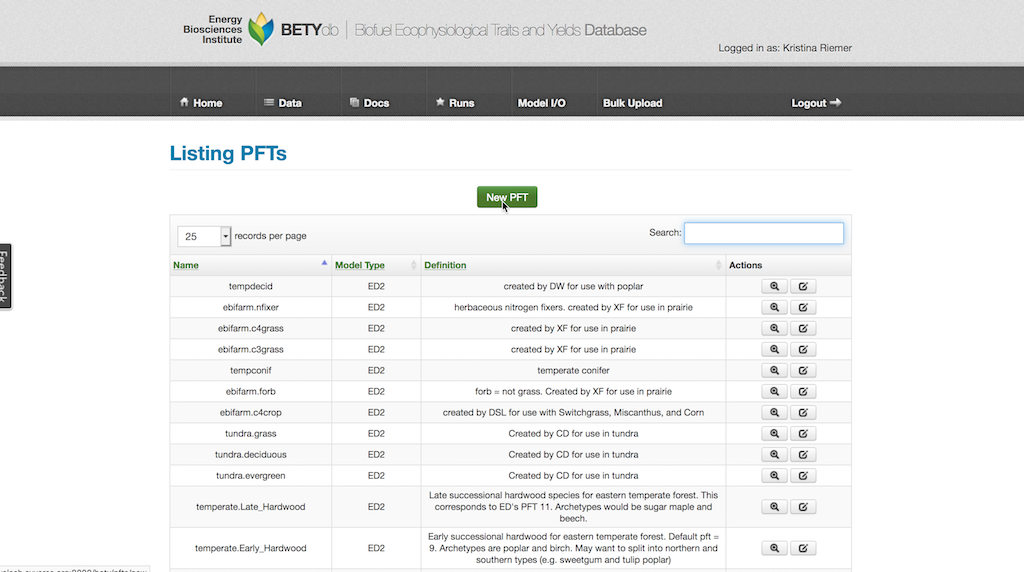
Give the PFT a descriptive name (e.g., temperate deciduous). PFTs are MODEL_TYPE specific, so choose your MODEL_TYPE from the pull down menu.
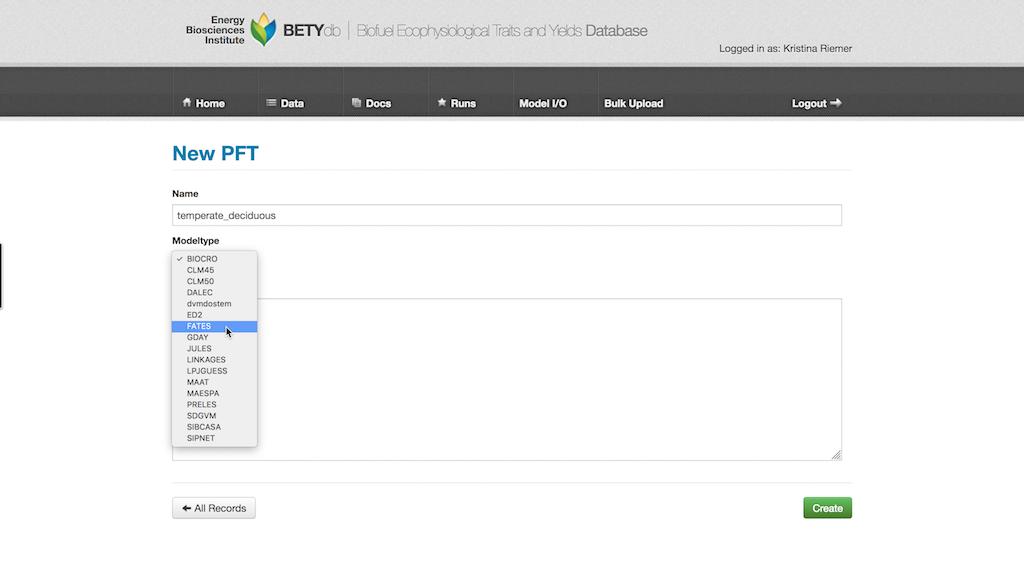
18.2.9.1 Species
Within PEcAn there are no predefined PFTs and user can create new PFTs very easily at whatever taxonomic level is most appropriate, from PFTs for individual species up to one PFT for all plants globally. To allow PEcAn to query its trait database for information about a PFT, you will want to associate species with the PFT record by choosing Edit and then “View Related Species”. Species can be searched for by common or scientific name and then added to a PFT using the +/- button.
18.2.9.2 Cultivars
You can also define PFTs whose members are cultivars instead of species. This is designed for analyses where you want to want to perform meta-analysis on within-species comparisons (e.g. cultivar evaluation in an agricultural model) but may be useful for other cases when you want to specify different priors for some member of a species. You cannot associate both species and cultivars with the same PFT, but the cultivars in a cultivar PFT may come from different species, potentially including all known cultivars from some of the species, if you wish to and have thought about how to interpret the results.
It is not yet possible to add a cultivar PFT through the BETYdb web interface. See this GithHub comment for an example of how to define one manually in PostgreSQL.
18.2.10 Adding Priors for Each Variable
In addition to adding species, a PFT is defined in PEcAn by the list of variables associated with the PFT. PEcAn takes a fundamentally Bayesian approach to representing model parameters, so variables are not entered as fixed constants but as prior probability distributions.
There are a wide variety of priors already defined in the PEcAn database that often range from very diffuse and generic to very informative priors for specific PFTs.
These pre-existing prior distributions can be added to a PFT. Navigate to the PFT from Data > PFTs and selecting the edit button in the Actions column for the chosen PFT.
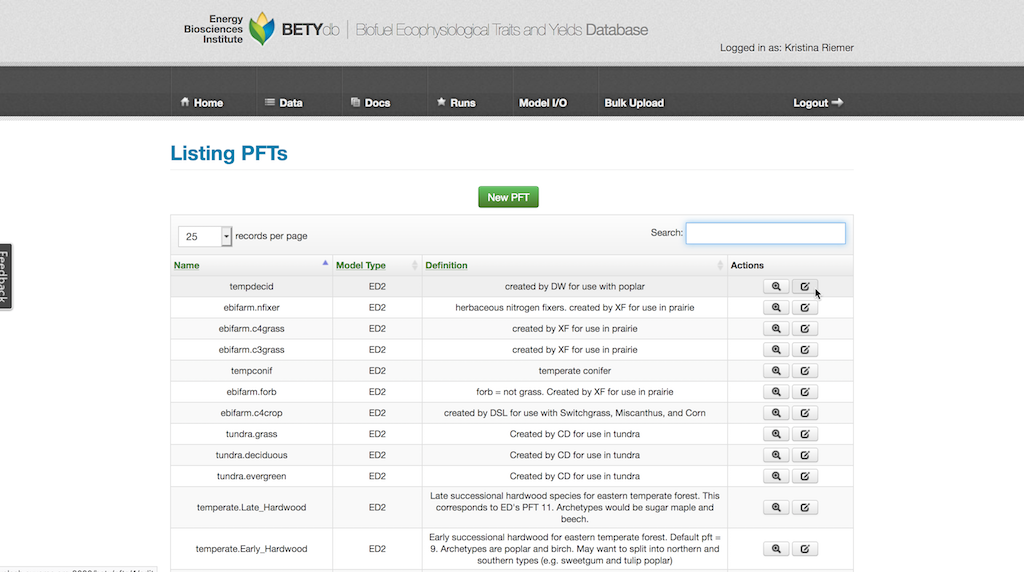
Click on “View Related Priors” button and search through the list for desired prior distributions. The list can be filtered by adding terms into the search box. Add a prior to the PFT by clicking on the far left button for the desired prior, changing it to an X.
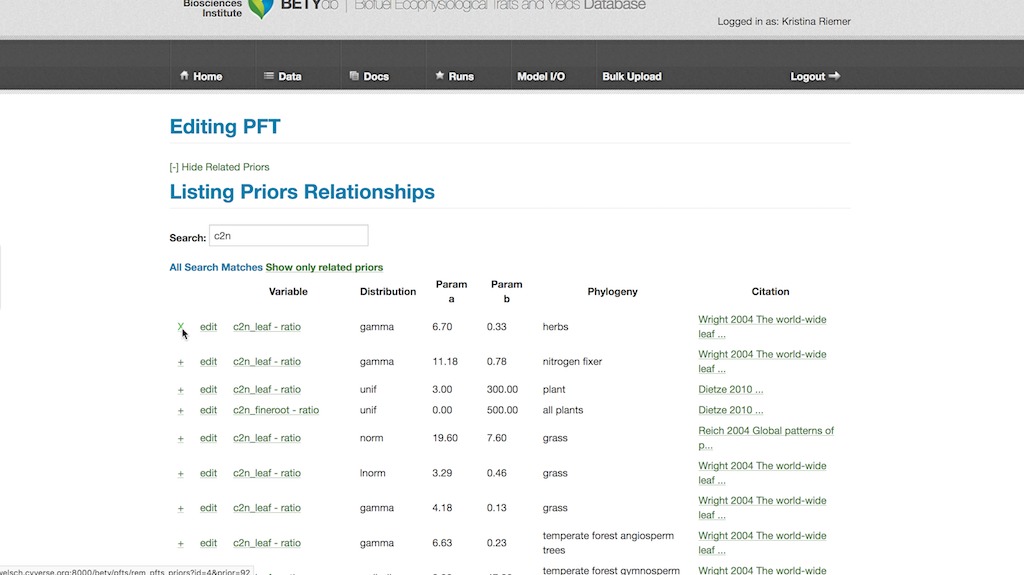
Save this by scrolling to the bottom of the PFT page and hitting the Update button.
18.2.10.1 Creating new prior distributions
A new prior distribution can be created for a pre-existing variable, if a more constrained or specific one is known.
- Select Data > Priors then “New Prior”
- In the Citation box, type in or select an existing reference that indicates how the prior was defined. There are a number of unpublished citations in current use that simply state the expert opinion of an individual
- Fill the Variable box by typing in part or all of a pre-existing variable’s name and selecting it
- The Phylogeny box allows one to specify what taxonomic grouping the prior is defined for, at it is important to note that this is just for reference and doesn’t have to be specified in any standard way nor does it have to be monophyletic (i.e. it can be a functional grouping)
- The prior distribution is defined by choosing an option from the drop-down Distribution box, and then specifying values for both Parameter a and Parameter b. The exact meaning of the two parameters depends on the distribution chosen. For example, for the Normal distribution a and b are the mean and standard deviation while for the Uniform they are the minimum and maximum. All parameters are defined based on their standard parameterization in the R language
- Specify the prior sample size in N if the prior is based on observed data (independent of data in the PEcAn database)
- When this is done, scroll down and hit the Create button
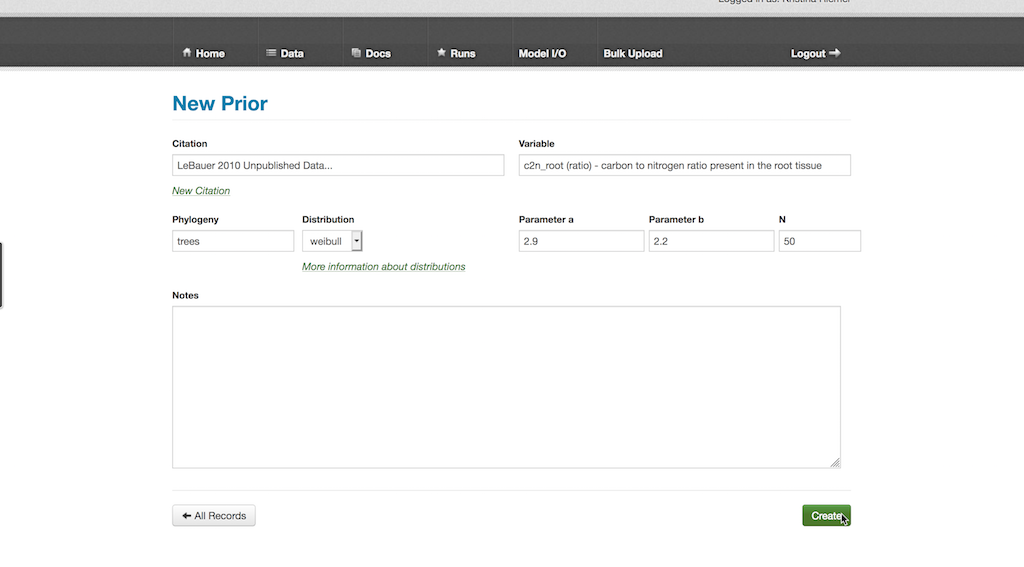
The new prior distribution can then be added a PFT as described in the “Adding Priors for Each Variable” section.
18.2.10.2 Creating new variables
It is important to note that the priors are defined for the variable name and units as specified in the Variables table. If the variable name or units is different within the model it is the responsibility of write.configs.MODEL function to handle name and unit conversions (see Interface Modules below). This can also include common but nonlinear transformations, such as converting SLA to LMA or changing the reference temperature for respiration rates.
To add a new variable, select Data > Variables and click the New Variable button. Fill in the Name field with the desired name for the variable and the units in the Units field. There are additional fields, such as Standard Units, Notes, and Description, that can be filled out if desired. When done, hit the Create button.
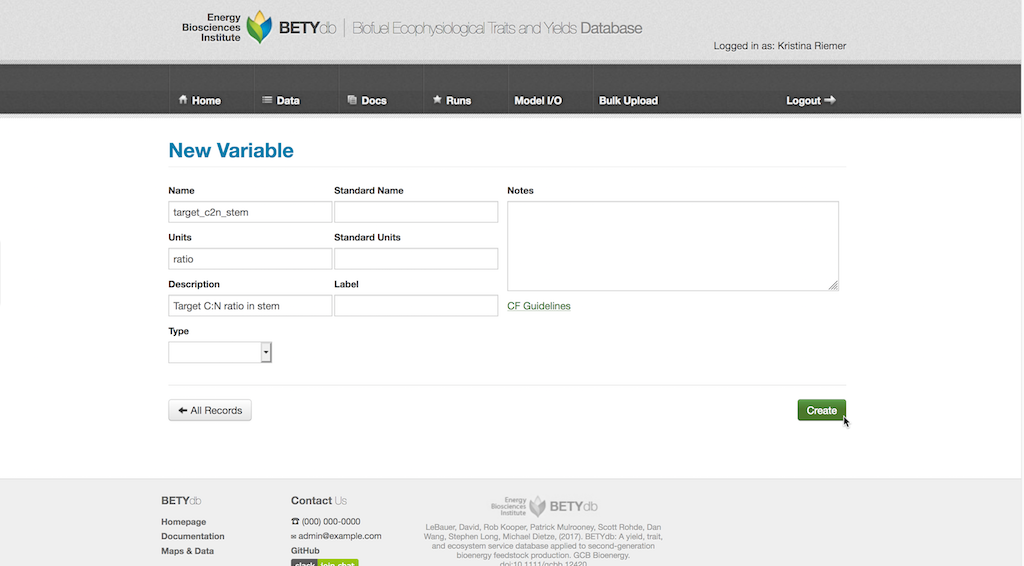
The new variable can be used to create a prior distribution for it as in the “Creating new prior distributions” section.