5 Adding Trait and Yield Data
5.1 The New Trait and New Yield Pages
5.1.1 Adding a Trait
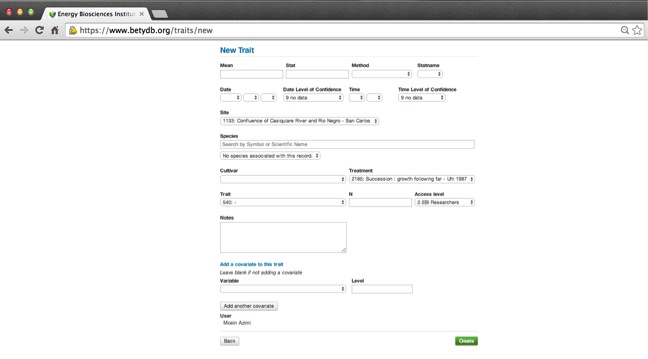
In general, a 'trait' is a phenotype (a characteristic that the plant exhibits). The traits that we are primarily interested in collecting data for are listed in Table 5.1. Before adding trait data, it is necessary to have the citation, treatments, and site information already entered. To add a new Trait, go to the new trait page.
| Variable | Units | Median (90%CI) or Range | Definition |
|---|---|---|---|
| Vcmax | \(\mu\)mol CO\(_2\) m\(^{-2}\) s\(^{-1}\) | \(44 (12, 125)\) | maximum rubisco carboxylation capacity |
| SLA | m\(^2\) kg\(^{-1}\) | \(15(4,27)\) | Specific Leaf Area area of leaf per unit mass of leaf |
| LMA | kg m\(^{-2}\) | \(0.09 (0.03, 0.33)\) | Leaf Mass Area (LMA = SLM = 1/SLA) mass of leaf per unit area of leaf |
| leafN | % | \(2.2(0.8, 17)\) | leaf percent nitrogen |
| c2n leaf | leaf C:N ratio | \(39(21,79)\) | use only if leafN not provided |
| leaf turnover rate | 1/year | \(0.28(0.03,1.0) \) | |
| Jmax | \(\mu\) mol photons m\(^{-2}\) s\(^{-1}\) | \(121(30, 262)\) | maximum rate of electron transport |
| stomatal slope | \(9(1, 20)\) | ||
| GS | stomatal conductance (= gs\(_{\textrm{max}}\)) | ||
| q* | 0.2--5 | ratio of fine root to leaf biomass | |
| **grasses* | ratio of root:leaf = below:above ground biomass | ||
| aboveground biomass | g m\(^{-2}\) or g plant\(^{-1}\) | ||
| root biomass | g m\(^{-2}\) or g plant\(^{-1}\) | ||
| **trees* | ratio of fine root:leaf biomass | ||
| leaf biomass | g m\(^{-2}\) or g plant\(^{-1}\) | ||
| fine root biomass (<2mm) | g m\(^{-2}\) or g plant\(^{-1}\) | ||
| root turnover rate | 1/year | 0.1--10 | rate of fine root loss (temperature dependent) year\(^{-1}\) |
| leaf width | mm | 22(5,102) | |
| growth respiration factor | % | 0--1 | proportion of daily carbon gain lost to growth respiration |
| R\(_{\textrm{dark}}\) | \(\mu\) mol CO\(_2\) m\(^{-2}\) s\(^{-1}\) | dark respiration | |
| quantum efficiency | % | 0--1 | efficiency of light conversion to carbon fixation, see Farqhuar model |
| dark respiration factor | % | 0--1 | converts Vm to leaf respiration |
| seedling mortality | % | 0--1 | proportion of seedlings that die |
| r fraction | % | 0--1 | fraction of storage to seed reproduction |
| root respiration rate* | CO\(_2\) kg\(^{-1}\) fine roots s\(^{-1}\) | 1--100 | rate of fine root respiration at reference soil temperature |
| f labile | % | 0--1 | fraction of litter that goes into the labile carbon pool |
| water conductance |
Presently, we are also using the Trait table to record ecosystem level measurements other than Yield. Such ecosystem level measurements can include leaf area index or net primary productivity, but are only collected when required for a particular project. Most of the fields in the Traits table are also used in the Yields table. Here is a list of the fields with a brief description, followed by more thorough explanations:
- Species: Search for species in the database using the search box; if species is not found, then the new species should be added to the database.
- Cultivar: Primarily used for crops.
- DateLOC: Date Level of confidence. See table 5.2 for values.
- TimeLOC: Time Level of confidence. See table 5.3 below for values.
- Mean: For yield,
meanis in units of tons per hectare per year (t/ha) - Stat name: The name of the statistical method used (usually one of SE, SD, MSE, CI, LSD, HSD, MSD). See Statistics for more details.
- Statistic: The value of the statistic associated with Stat name.
- N: Always record N if provided. N is the number of experimental replicates, often referred to as the sample size; N represents the number of independent units within each treatment: in a field setting, this is often the number of plots in each treatment, but in a greenhouse, growth chamber, or pot-study this may be the number of chambers, pots, or individual plants. Sometimes this value is not clearly stated.
5.1.1.1 Uncertainty in Date or Time
DateLOC
The date level of confidence (DateLOC, Table ) provides an indication of how accurately the date associated with the trait or yield observation is known. It provides the values that should be entered in this field. If the event occurred at a level of precision not defined by an integer in this table, then use fractions. For example, we commonly use 5.5 to indicate a one week level of precision. If the exact year is not known, but the time of year is, then use 91 to 97, with the second digit to indicate the information known within the year.
| Dateloc | Definition |
|---|---|
| 9 | no data |
| 8 | year |
| 7 | season |
| 6 | month |
| 5 | day |
| 95 | unknown year, known day |
| 96 | unknown year, known month |
| ...etc |
TimeLOC
The time level of confidence (TimeLOC) provides an indication of how accurately the time associated with the trait or yield observation is known. It provides the values that should be entered in this field.
| Timeloc | Definition |
|---|---|
| 9 | no data |
| 4 | time of day i.e. morning, afternoon |
| 3 | hour |
| 2 | minute |
| 1 | second |
5.1.1.2 Statistics
Our goal is to record statistics that can be used to estimate standard deviation or standard error (https://www.authorea.com/users/5574/articles/6811/). Many different methods can be used to summarize data, and this is reflected in the diversity of statistics that are reported. An overview of these methods is given in a description below.
Where available, direct estimates of variance are preferred, including Standard Error (SE), sample Standard Deviation (SD), or Mean Squared Error (MSE). SE is usually presented in the format of mean (±SE). MSE is usually presented in a table. When extracting SE or SD from a figure, measure from the mean to the upper or lower bound. This is different than confidence intervals and range statistics (described below), for which the entire range is collected.
If MSE, SD, or SE are not provided, it is possible that LSD, MSD, HSD, or CI will be provided. These are range statistics and the most frequently found range statistics include a Confidence Interval (95%CI), Fisher’s Least Significant Difference (LSD), Tukey’s Honestly Significant Difference (HSD), and Minimum Significant Difference (MSD). Fundamentally, these methods calculate a range that indicates whether two means are different or not, and this range uses different approaches to penalize multiple comparisons. The important point is that these are ranges and that we record the entire range.
Another type of statistic is a “test statistic”; most frequently there will be an F-value that can be useful, but this should not be recorded if MSE is available. Only if there is no other information available should you record the P-value.
5.1.2 Adding a Yield
The protocol for entering yield data is identical to entering data for a trait, with a few exceptions:
There are no covariates associated with yield data
Yield data is always the dry harvestable biomass; if necessary, moisture content can be added as a trait
Yield is equivalent to aboveground biomass on a per-area basis, and has units of Mg ha\(^{-1}\) y\(^{-1}\).
5.1.3 Adding a Covariate
Covariates are required for many of the traits. Covariates generally indicate the environmental conditions under which a measurement was made. Without covariate information, the trait data will have limited value.
A complete list of required covariates can be found in Table 5.4. For all respiration rates and photosynthetic parameters, temperature is recorded as a covariate. Soil moisture, humidity, and other such variables that were measured at the time of the measurement may be required in order to standardize across studies.
When root data is recorded, the root size class needs to be entered as a covariate. The term ’fine root’ often refers to the <2mm size class, and in this case, the covariate root_maximum_diameter would be set to 2. If the size class is a range, then the root_minimum_diameter can also be used.
Table 5.4: Traits with required covariates
A list of traits and the covariates that must be recorded along with the trait value in order to be converted to a constant scale from across studies.
notes:
Stomatal conductance (gs) is only useful when reported in conjunction with other photosynthetic data, such as Amax. Specifically, if we have Amax and gs, then estimation of Vcmax only covaries with dark_respiration_factor and atmospheric CO2 concentration.
We also now have information to help constrain stomatal_slope. If we have Amax but not gs, then our estimate of Vcmax will covary with: dark_respiration_factor, CO2, stomatal_slope, cuticular_conductance, and vapor-pressure deficit VPD (which is more difficult to estimate than CO2, but still possible given lat, lon, and date). Most important, there will be a strong covariance between Vcmax and stomatal_slope.
| Variable | Required Covariates | Optional Covariates |
|---|---|---|
| vcmax | temperature (leafT or airT) | irradiance |
| any leaf measurement | canopy_height or canopy_layer | |
| root_respiration_rate | temperature (rootT or soilT) | soil moisture |
| root_diameter_max | root size class (usually 2mm) | |
| any respiration | temperature | |
| root biomass | min. size cutoff, max. size cutoff | |
| root, soil | depth (cm) | used for max and min depths of soil, if only one value, assume min depth = 0; negative values indicate above ground |
| gs (stomatal conductance) | Amax | see notes in caption |
| stomatal_slope (m) | humidity, temperature | specific humidity, assume leaf T = air T |
| SLA | canopy_level |
5.2 Bulk Upload
5.2.1 Overview
There are three phases for a basic bulk upload of data:
Enter meta-data pertaining to your data set (new sites, species, cultivars, citations, or treatments).
Create a CSV file of the appropriate form that contains your data.
You may use one of the following sample headings to get started.
Heading for yield data with citation specified by author, year, and title.
citation_author,citation_year,citation_title,cultivar,species,site,treatment,date,yield,n,SE,notes,access_level
If your data includes standard error and cultivar information and you do not plan to specify any of the required information interactively, you will be able to use this template “as-is”. Otherwise, you will need to delete one or more columns:
If your data has no standard error information, delete both the
SEand thencolumn.If your data set has a single uniform value for the site, species, cultivar, treatment, access_level, or date, then these values may be entered interactively through the web interface; in this case you should delete the corresponding column(s) from the template.
Note that cultivar information can’t be specified interactively unless the species information is as well (and vice versa); delete the
cultivarcolumn if and only if you either have no cultivar information or you are specifying both the species and the cultivar interactively.
Heading for yield data with citation specified by DOI.
citation_doi,cultivar,species,site,treatment,date,yield,n,SE,notes,access_level
Use this template if you are uploading yield data and you wish to specify the citation in the file by its Digital Object Identifier (DOI).
Again, if you do not have data for all of the columns listed in the template, or if you plan to specify some of the data interactively, you will have to delete one or more columns.
You may also use this template if all of the data in your data set pertains to a single citation and you wish to specify that citation interactively. In this case, you must delete the
citation_doicolumn.
Heading for trait data with citation specified by author, year, and title.
citation_author,citation_year,citation_title,cultivar,species,site,treatment,date,entity,[trait variable 1],[trait variable 2],[trait variable n],[covariate 1],[covariate 2],[covariate n],n,SE,notes,access_level
Use this template if you are uploading trait data and you wish to specify the citation in the file by author, year, and title.
This template must be modified before it can be used. In particular, the column headings
[trait variable 1]…[trait variable n]must be replaced by actual variable names that exactly match names of variables in the database. The number of these trait variable columns may need to be increased or decreased to accommodate the data set.Some trait variables allow or even require corresponding covariate information to be included. Again, the column headings
[covariate 1]…[covariate n]must be changed to actual covariate variable names, and the number of these columns may need to be increased or decreased to match the available information. For a list of recognized trait variable names that are treated as covariates, visit the trait variable/covariates list at https://www.betydb.org/trait_covariate_associations or the corresponding page of the site to which you are uploading your data.As with the yield data templates, delete columns for attributes that have a uniform value for the data set and that you plan to specify interactively.
Trait data files may also include an
entitycolumn if you wish to use named entities to group associated traits. Delete this column heading from the template if you simply want to use anonymous automatically-created entities. For more detailed information about entities, see Optional data below.
Heading for trait data with citation specified DOI.
citation_doi,cultivar,species,site,treatment,date,entity,[trait variable 1],[trait variable 2],[trait variable n],[covariate 1],[covariate 2],[covariate n],n,SE,notes,access_level
As with the corresponding yield data template, use this template if you are uploading trait data and you wish to specify the citation in the file by DOI or if you plan to specify the citation interactively (in which case delete the
citation_doicolumn). Again, this template must be modified before it can be used.Use the Bulk Upload Wizard in the BETYdb web interface to upload your data set and insert it into the database.
5.2.2 Detailed CSV Data File Specifications
In what follows, the term “field” always refers either to a column name used in the heading of the uploaded CSV file or to an entry in that column in some particular row of the file. On the other hand, and the term “column” may either refer to a column of data in the uploaded CSV file or to an attribute of a trait or yield datum in the traits or yields table of the database.
5.2.2.1 General Considerations
Although the comma-separated value (CSV) file format is not fully standardized, here are some general guidelines:
The top line of the file should contain a comma-separated list of column headings. Any column heading not recognized as being significant (see below) will be ignored, as will the data in subsequent rows falling under that heading.
Lines (or rows) after the first contain data. Data items are separated by commas, and in general there will be one data item for each column heading; the nth data item in a row "belongs" to the nth heading in the top line.1
There can be no extraneous blank lines, even at the end of the file.
Extraneous space before or after a comma or at the beginning or end of a row is discarded.
To include a comma as part of a data item, the data item must be quoted with double-quotes so that the comma is not interpreted as a data-value separator. There should be no extraneous space between a double-quoted item and the preceding and following commas or beginning or end of the line.
To include a literal double quote as part of a quoted item, use two consecutive double quotes.
5.2.2.2 Required data file fields
For yields uploads, the only required field is a
yieldcolumn.For trait uploads, there must be at least one column whose label exactly matches the variable name for the trait value being specified. (Leading and trailing spaces are permitted, but letter case must exactly match the name of the variable specified in the database.) If this trait variable has any required covariates, columns for these covariates must be included. Again, visit https://www.betydb.org/trait_covariate_associations (or the corresponding page for the site you are uploading to) to see which traits require covariates.
5.2.2.3 Information that is required but that may be specified interactively for the entire dataset.
Attributes of data items may be specified interactively only if there is a single uniform value that pertains to the whole data set contained in the upload file.
Information that references existing database entries
As mentioned, the first step in doing a bulk upload is to ensure that there are existing items already in the database for each attribute value of each item in the upload set. There are defined rules as to how to refer to these existing attributes in your upload file. For example, a species must be referred to by its scientific name even though there are other attributes of the species items stored in the species table that might uniquely identify a particular species (the AcceptedSymbol attribute, for example).
In these sections, we specify, for each attribute, how to refer to the attribute value, whether or not the attribute can be specified interactively or must be specified in the data file, and what criteria are used to match values specified in the data file with existing table entries in the database.
Citation
If the entire dataset pertains to a single citation, that citation may be specified interactively by choosing a citation on the citations page instead of including citation information in the CSV file.
Otherwise, specify the citations in the CSV file, either by DOI or by author, year, and title.
If a DOI is available for all citations in the data set, the citation corresponding to each row may be specified in a
citation_doicolumn. In this case, thecitation_author,citation_year, andcitation_titlecolumns must not be in the column heading list.2 Each value in thecitation_doicolumn must exactly match thedoiattribute of some row in thecitationstable except that letter case and leading and trailing spaces are ignored.Conversely, if a DOI is not available for all citations in the data set, or if it is preferred to specify the citation by author, year, and title, then the
citation_doicolumn should not be included and the columnscitation_author,citation_year, andcitation_titlemust all be present.3
Site
If all of the data in the data set pertains to a single site, that site may be specified interactively.
Otherwise, a
sitecolumn is required. Each value in the column must match an existingsitenamecolumn value in thesitestable of the database. (Letter case, leading and trailing spaces, and extra internal spaces are ignored when searching for a match.)
Species
If all of the data in the data set pertains to a single species, that species may be specified interactively.
Otherwise, the
speciescolumn is required. Each value must match an existingscientificnamecolumn value in thespeciestable of the database. (Again, letter case, leading and trailing spaces, and extra internal spaces are ignored when searching for a match.)
Treatment
If a single treatment and a single citation applies to all of the data in the data set, the treatment may be specified interactively provided that the citation is specified interactively as well.
Otherwise, a
treatmentcolumn is required. The value must match an existingnamecolumn value in thetreatmentstable of the database; moreover, this matching treatment must be consistent with the specified citation. (Again, letter case, leading and trailing spaces, and extra internal spaces are ignored when searching for a match.)
Other information that may be specified interactively
Date
If a single date (with or without time of day) applies to all of the data in the data set, the date may be specified interactively.
Otherwise, if there are multiple dates (with or without times) associated with the data, a
local_datetimecolumn is required.4Values in the
local_datetimecolumn or in thedatetext box in the Global Values page may be in one of two forms:Date values without time of day must be in the form
YYYY-MM-DD. For example, July 25, 2003 must be entered as “2003-07-25”.Values that include the time of day must be in the form
YYYY-MM-DD HH:MM:SSorYYYY-MM-DDTHH:MM:SS. (Here,Tis simply the literal letterT.) For example, 11 a.m. on November 11, 1918 could be specified either as "1918-11-11 11:00:00" or as "1918-11-11T11:00:00". Note that the yields table stores only the date and so any time-of-day information given in a yields data file or specified interactively will be ignored..
Note that the date and time should always be the date and time at the site where the trait measurement was made.5 When uploading trait data, each site associated with the data file must be associated with a valid time zone!67
Rounding
The amount of rounding for numerical data can only be specified interactively. Any value from 1 to 4 significant digits may be chosen. The amount of rounding for the standard error SE (if present) may be specified separately from the amount of rounding for yield and for trait variables and their covariates.
By default, all numerical measurements are rounded to three significant digits. For example, with this default in place, 999.1 will be rounded to 999 and 1001.1 will be rounded to 1000.
Standard error values (if given) are by default rounded to two significant digits.
5.2.2.4 Numerical Data
Numerical data is never specified interactively since typically each measurement will have a different numerical value so there will never be a uniform value for all data in the data set.
Data for Yields
Yield
Every yield data upload file must have a
yieldcolumn. The data in this column must always be a parsable non-negative number and must never be blank. Scientific notation is not currently supported. As noted above, the number given in the file is subject to rounding before being inserted into the database.Sample Size
An
ncolumn is required if and only if anSEcolumn is included. The value must always be an integer greater than 1.Standard Error
An
SEcolumn is required if and only if anncolumn is included; this datum will be inserted into thestatcolumn of theyieldstable, and thestatnamecolumn value will be set to “SE”. As with the yield column, the data in this column must never be blank, and values can't be in scientific notation.8
Data for Traits
Trait variable values
Every trait data upload file must have at least one column whose heading matches the name of some recognized trait variable. A list of recognized trait variables is listed on the BETYdb web site. If multiple trait variable columns are used, each row in the CSV file will produce one row in the
traitstable for each trait variable column having a non-blank entry.9 (These resulting rows will be effectively grouped by assigning them a unique entity id. Said another way, there is a one-to-one correspondence between rows in the CSV file and resultant rows in theentitiestable, the table that keeps track of this grouping.) As with yield numbers, the data in this column must always be a parsable number and is subject to rounding before being inserted into the database. In addition, it must conform to any range restrictions on the value of the variable.The template for traits uploads provides dummy column headings
[trait variable 1],[trait variable 2], etc., which must be changed to actual variable names before data can be uploaded.
Covariate values
If any of the included trait variables has a required covariate, there must be a column corresponding to that covariate.
For any of the included trait variables that has an optional covariate, a column corresponding to that covariate may be included.
As with trait variable columns, entries in a covariate variable column may be left blank.10
The template for traits uploads provides dummy column headings
[covariate 1],[covariate 2], etc., which must be changed to actual variable names before data can be uploaded.
Sample Size and Standard Error
An
SEcolumn is required if and only if anncolumn is included; this datum will be inserted into thestatcolumn of thetraitstable, and thestatnamecolumn value will be set to “SE”. *Note that if you have more than one trait variable column, thenandSEcolumns are not allowed. This is because there is currently no way to use different standard error values for different trait variables, and it is unlikely that different variables would share the same value for the standard error.11Note that even if
nandSEvalues are specified, thenandSEvalues for any associated covariates will be set to NULL: There is currently no way to specify standard error values for covariates in a bulk upload file.Again, values of
nmust be at least 2, and columns fornandSEmust both be present or both be absent.
5.2.2.5 Optional data
Sample Size and Standard Error
As noted above, these are both optional, but if one of these is included, the other must be included as well. In other words, the column heading list must include both of
nandSEor neither. Note that ifnandSEare not included in the uploaded CSV file, the value of thencolumn of the traits or yields table will default to 1 and thestatandstatnamecolumn values will default to NULL and the empty string, respectively.Cultivar
If a uniform value for the species is provided interactively when uploading the data set, the cultivar may be specified this way as well, provided that it also has a uniform value for the whole data set.
Otherwise, to include cultivar information in the upload file, both a
speciesand acultivarcolumn must be included. The values in thecultivarcolumn are allowed to be blank (in which case a value of NULL is inserted into thecultivar_idcolumn for the given row); but if provided, the value must match the value of thenamecolumn in some row of thecultivarstable; moreover, this row must be a row associated with the species corresponding to the value given in thespeciescolumn. Again, matching is case insensitive, and leading, trailing, and excess internal whitespace is ignored.
Notes
To include notes, use a
notescolumn. There is no restriction on what can be included in this column, but leading and trailing space will be stripped before insertion into the database. Non-ascii characters entered in the file in UTF-8 encoding are allowed. If there is nonotescolumn, each row inserted into thetraitsoryieldstable will use the empty string as the value for thenotescolumn.Entities
Entities are a way of grouping related traits. For example, trait measurements made at the same time on the same plant group, plant, or plant part are intrinsically related. To show this in the database, they may be given a common value for the
entity_idattribute.The Bulk Upload Wizard handles entities as follows:
The data file is for yields. In this case, no entities are created, and all yield rows created will have NULL as their
entity_idattribute.The data file is for traits and has no "entity" column but has at least two trait-variable columns. In this case, what happens for a given row will depend on whether that row has any non-blank trait-variable column values.
If the row has at least one non-empty trait-variable column value, a new, anonymous entity will be created and each trait created for the row will be assigned the new entity.
If all trait-variable column values in the row are blank, no entities or traits are created for that row.
The data file is for traits, has no "entity" column, and has only one trait-variable column. In this case, no entities are created, and all trait rows created will have NULL as their
entity_idattribute.The data file is for traits and does have an "entity" column. In this case, what happens for a given row will depend on whether the "entity" column and the trait-variable columns are blank.
If the "entity" column is not blank and the name given there matches the name of an existing entity, the new traits created for the row will be assigned to that entity.
If the "entity" column is not blank but does not match the name of any existing entity, and if at least one trait-variable column value is non-blank, a new entity will be created whose name is the value specified in the "entity" column, and new traits created for the row will be assigned to this new entity.
If the "entity" column is not blank but all trait-variable column values are blank, no new entity will be created whether or not the value in the "entity" column matches the name of an existing entity.
Rows in which the "entity" column is blank are treated as in the case that the entity column does not exist: that is, anonymous entities are only created if there are at least two trait-variable columns and then only for rows for which at least one trait-variable column value is non-blank.
Methods
A method may be specified for each trait variable in the data file. Methods must be specified interactively on the Upload Options and Global Values page. This means of course that for each file, each trait variable must be assigned uniformly to the same method; trait values in different rows can not be assigned different methods.
5.2.3 Using the Bulk Upload Wizard
Once a CSV data file has been composed, you are ready to start an upload using the Bulk Upload Wizard.
The Bulk Upload Wizard is a sequence of pages designed to guide the user through the process of uploading a data file and having its data added to the database. Here is a step-be-step guide to using the wizard. This guide assumes you have already composed a data file, and that it is accessible in the file system of the computer you are using to access the wizard.
Click the Bulk Upload menu item at the top of any BETYdb page.
On the "New Bulk Upload" page, click the "Choose File" button. In the file chooser that comes up, find the file you want to upload and double-click on it.
Click the "Upload and validate file" button.
If you do not have citation information in your file and you have not chosen a citation to use in your browser session, you will be taken to the "Choose a Citation" page. Otherwise, skip the next steps.
Type into the auto-completion citation text box and choose from among the list of citations that come up. Note that the list of citations will be filtered by trying to match the author, year, title, or DOI against the string of characters you type into the box.
Once you have chosen a citation, click the "View Validation Results" button.
View the validation results. This page will display your data file and will highlight any errors or possible errors. For details on potential problems, see the Troubleshooting section.
If there are no errors to be dealt with, click the link on the upper-right to go on to the "Specify Upload Options and Global Values" page.
The "Specify Upload Options and Global Values" page allows for specifying global value that apply to the whole data set.
At a minimum—if all meta-data which can be specified in the data file is specified there—one will be presented with the option to specify the amount of rounding of yield values.
If the file contains standard error information, one will also be presented with the option to specify the amount of rounding of standard error values.
If any of the attributes "site", "species", "treatment", "access_level", or "date" was not specified in the data file, one is required to enter values for them here.
If the species is being specified interactively, one will be presented with the option to specify the cultivar interactively as well.
For trait uploads, one will be presented with the option to specify a method for each trait variable in the file.
Once you have specified all required values and any optional value you with to specify, click "Confirm Data".
The "Verify Upload Specifications and Data-Set References" will present a summary both of information specified in the data file and information you specified interactively.
If the upload file is a trait file, each trait variable will be listed along with any associated covariates specified in the data file and any methods specified on the previous page.
If the data access level was specified interactively, the value chosen will be displayed.
The amount of rounding chosen for yield or trait values and (if applicable) for standard error values will be displayed.
All reference data (meta-data) for the data set will be listed:
All citations referenced will be listed, with the author, title, year, DOI, and journal (if specified).
If any named entities were specified in the file, they will be listed along with their parent entity (if any) and notes (if any).
The names of all associated sites will be listed, along with their city, state, country, latitude and longitude, and (if specified) the soil type and soil notes.
Each associated species will be listed, including (if specified) the full scientific name, the genus, the common name, and the accepted symbol.
If cultivars were specified, their names will be listed along with their ecotype and associated species.
Names of all treatments specified will be listed, together with (if specified) their definition, whether or not they are a control, and the author of the associated citation.
Once the summary has been checked over for any errors or omissions, click the "Insert Data" button if all is OK. If the upload was successful, you will be returned to the start page of the wizard and a success message will be displayed.
5.2.4 Troubleshooting
[to-do]
In some cases, it is allowable for data in some columns to be missing from some rows. In this case, there will be two consecutive commas with only zero or more white-space characters in between, or, in the case of the first or last columns, there may be only whitespace before the first or after the last comma of the row. If items are omitted from the end of a line, the trailing commas may be omitted as well.↩
If such information is already included in the data set and you want to keep such columns for purely informational purposes, the string
-ignoremay be appended to each of these headings. (In fact, renaming the columns to any non-recognized heading name will do.)One might want to do this, for example, to keep a visual record of the author, year, and title even when it is the citation DOI that is being used to determine how the data will associated with a citation in the database.↩
Again, if some DOI information is already included and you wish to retain it for purely informational purposes, you could append
-ignoreto thecitation_doiheading.↩This column can also be called
date, but using this as the column heading is deprecated:local-datetimemore clearly emphasizes that the dates or dates-and-time specified in this column are interpreted as being the date/time in the time zone of the site where the trait measurement was made.↩The
dateloc(Date Level of Confidence) value is always set automatically to 5 ("day"). Thetimeloc(Time Level of Confidence) value is automatically set to 1 ("second") if the time of day is given and 9 ("no data") otherwise.↩Currently, there is not way to specify a site timezone via the BETYdb web application. One must manipulate the database directly.↩
The rule is slightly different for a site that is specified interactively: If a time zone has been specified for the site, it must still be a valid one. But it isn't an error to specify a site whose time zone attribute is null. In this case, the date or date-and-time values specified will be interpreted as being in UTC.↩
Values should be non-negative, but this isn't currently enforced.
It is conceivable that one might have a data set in which some rows have SE values and some do not. The Bulk Upload Wizard doesn't currently handle such a case, and such a data set would have to be separated into two files: one with columns for
nandSE, and one without.↩Unlike yield data files, entries in trait or covariate variable columns of a trait data file are allowed to be blank. In this case, no trait is inserted into the database for that variable of that row of the data file.↩
There is a small loophole in the way required covariates are enforced. While it is true that if a column for a trait having a required covariate is included, there must also be a column for that covariate, it is not flagged as an error if certain rows leave that covariate column blank.↩
Eventually, we may enable associating differing values of
nandSEto different trait variables and covariates. In this case, we might add columns[trait variable 1] nand[trait variable 1] SE, etc. or[covariate 1] nand[covariate 1] SE, prefixing the usual column heading with a variable name to indicate which variable the sample size and standard error value is to be associated with.↩